Ebola Data Loader¶
In this notebook, we’ll format data from The World Health Organization for future analysis
%pylab inline
import pandas as pd
import re
Populating the interactive namespace from numpy and matplotlib
#read in the raw data
rawdata = pd.read_csv('Ebola_in_SL_Raw_WHO_Data.csv')
rawdata.iloc[1]
COUNTRY (CODE) SLE
COUNTRY (DISPLAY) Sierra Leone
COUNTRY (URL) NaN
EBOLA_MEASURE (CODE) CASES
EBOLA_MEASURE (DISPLAY) Number of cases
EBOLA_MEASURE (URL) NaN
CASE_DEFINITION (CODE) CONFIRMED
CASE_DEFINITION (DISPLAY) Confirmed
CASE_DEFINITION (URL) NaN
EBOLA_DATA_SOURCE (CODE) PATIENTDB
EBOLA_DATA_SOURCE (DISPLAY) Patient database
EBOLA_DATA_SOURCE (URL) NaN
EPI_WEEK (CODE) 2015-W07
EPI_WEEK (DISPLAY) 09 to 15 February 2015
EPI_WEEK (URL) NaN
INDICATOR_TYPE (CODE) SITREP_NEW
INDICATOR_TYPE (DISPLAY) New
INDICATOR_TYPE (URL) NaN
DATAPACKAGEID (CODE) 2015-04-22
DATAPACKAGEID (DISPLAY) Data package 2015-04-22
DATAPACKAGEID (URL) NaN
Display Value 92.0
Numeric 92.0
Low NaN
High NaN
Comments NaN
Name: 1, dtype: object
#parse the dates column
import dateutil
def parsedate(week_string):
end_date_str = re.split(' to ', week_string)[1]
return(dateutil.parser.parse(end_date_str))
rawdata['End Date'] = rawdata['EPI_WEEK (DISPLAY)'].apply(parsedate)
rawdata.head()
| COUNTRY (CODE) | COUNTRY (DISPLAY) | COUNTRY (URL) | EBOLA_MEASURE (CODE) | EBOLA_MEASURE (DISPLAY) | EBOLA_MEASURE (URL) | CASE_DEFINITION (CODE) | CASE_DEFINITION (DISPLAY) | CASE_DEFINITION (URL) | EBOLA_DATA_SOURCE (CODE) | ... | INDICATOR_TYPE (URL) | DATAPACKAGEID (CODE) | DATAPACKAGEID (DISPLAY) | DATAPACKAGEID (URL) | Display Value | Numeric | Low | High | Comments | End Date | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | SLE | Sierra Leone | NaN | CASES | Number of cases | NaN | CONFIRMED | Confirmed | NaN | SITREP | ... | NaN | 2015-04-22 | Data package 2015-04-22 | NaN | NaN | NaN | NaN | NaN | NaN | 2014-02-23 |
| 1 | SLE | Sierra Leone | NaN | CASES | Number of cases | NaN | CONFIRMED | Confirmed | NaN | PATIENTDB | ... | NaN | 2015-04-22 | Data package 2015-04-22 | NaN | 92.0 | 92.0 | NaN | NaN | NaN | 2015-02-15 |
| 2 | SLE | Sierra Leone | NaN | CASES | Number of cases | NaN | CONFIRMED | Confirmed | NaN | PATIENTDB | ... | NaN | 2015-04-22 | Data package 2015-04-22 | NaN | 455.0 | 455.0 | NaN | NaN | NaN | 2014-10-19 |
| 3 | SLE | Sierra Leone | NaN | CASES | Number of cases | NaN | CONFIRMED | Confirmed | NaN | SITREP | ... | NaN | 2015-04-22 | Data package 2015-04-22 | NaN | 63.0 | 63.0 | NaN | NaN | NaN | 2015-02-22 |
| 4 | SLE | Sierra Leone | NaN | CASES | Number of cases | NaN | CONFIRMED | Confirmed | NaN | SITREP | ... | NaN | 2015-04-22 | Data package 2015-04-22 | NaN | 80.0 | 80.0 | NaN | NaN | NaN | 2015-02-01 |
5 rows × 27 columns
data = rawdata[rawdata['EBOLA_DATA_SOURCE (CODE)']=='PATIENTDB']
data = data[['End Date','Numeric']]
data.sort_values('End Date', inplace=True)
data.dropna(inplace=True)
data['Timedelta'] = data['End Date']-data['End Date'].iloc[0]
data['Weeks'] = data['Timedelta'].apply(lambda a: pd.Timedelta(a).days/7)
data.set_index('Weeks', inplace=True)
data = data[['Numeric']]
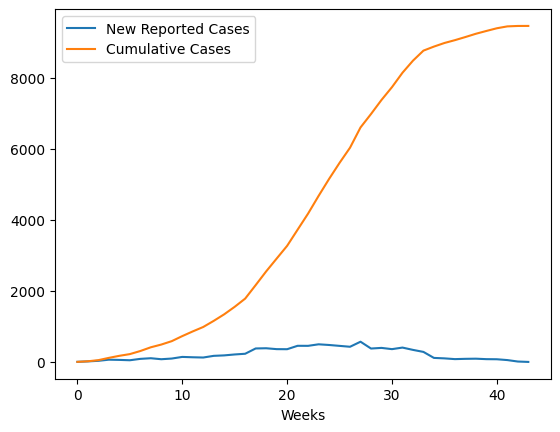
data.columns=['New Reported Cases']
data['Cumulative Cases'] = data['New Reported Cases'].cumsum()
data.plot()
<AxesSubplot:xlabel='Weeks'>
data.to_csv('Ebola_in_SL_Data.csv')